This chapter provides guidance and procedures used for characterization of biotechnology-derived articles by polyacrylamide gel electrophoresis. This chapter is harmonized with the corresponding chapter in JP and EP. Other characterization tests, also harmonized, are shown in Biotechnology-Derived Articles—Amino Acid Analysis  1052
1052 , Biotechnology-Derived Articles—Capillary Electrophoresis
, Biotechnology-Derived Articles—Capillary Electrophoresis  1053
1053 , Biotechnology-Derived Articles—Isoelectric Focusing
, Biotechnology-Derived Articles—Isoelectric Focusing  1054
1054 , Biotechnology-Derived Articles—Peptide Mapping
, Biotechnology-Derived Articles—Peptide Mapping  1055
1055 , and Biotechnology-Derived Articles—Total Protein Assay
, and Biotechnology-Derived Articles—Total Protein Assay  1057
1057 .
.
INTRODUCTION
Polyacrylamide gel electrophoresis (PAGE) is used for the qualitative characterization of proteins in biological preparations, for control of purity, and for quantitative determinations. This procedure is limited to the analysis of proteins with a weight range of 14,000 to 100,000 Da. It is possible to extend the weight range of an electrophoresis gel by various techniques (e.g., gradient gels or particular buffer systems), but such techniques are not discussed in this chapter. Analytical gel electrophoresis is an appropriate method with which to identify and to assess the homogeneity of proteins in drug substances. These methods are routinely used for the estimation of protein subunit molecular weights and for the determination of the subunit compositions of purified proteins.
Ready-to-use gels and reagents are commercially available and can be used instead of those described in this chapter, provided that they give equivalent results and that they meet the validation requirements.
GENERAL PRINCIPLE OF ELECTROPHORESIS
Under the influence of an electrical field, charged particles migrate in the direction of the electrode bearing the opposite polarity. In gel electrophoresis, the movements of the particles are retarded by interactions with the surrounding gel matrix, which acts as a molecular sieve. The opposing interactions of the electrical force and molecular sieving result in differential migration rates according to the sizes, shapes, and charges of particles. Because of their different physicochemical properties, different macromolecules of a mixture migrate at different speeds during electrophoresis and thus are separated into discrete fractions. Electrophoretic separations can be conducted in systems without support phases (e.g., free solution separation in capillary electrophoresis) and in stabilizing media, such as thin-layer plates, films, or gels.
CHARACTERISTICS OF POLYACRYLAMIDE GELS FOR PROTEIN ELECTROPHORESIS
The sieving properties of polyacrylamide gels are established by the three-dimensional network of fibers and pores that is formed as the bifunctional bisacrylamide cross-links adjacent to polyacrylamide chains. Polymerization is catalyzed by a free-radical-generating system composed of ammonium persulfate and N,N,N¢,N¢-tetramethylethylenediamine (TEMED).
As the acrylamide concentration of a gel increases, its effective pore size decreases. The effective pore size of a gel is operationally defined by its sieving properties, that is, by the resistance it imparts to the migration of macromolecules. There are limits to the acrylamide concentrations that can be used. At high acrylamide concentrations, gels break much more easily and are difficult to handle. As the pore size of a gel decreases, the migration rate of a protein through the gel decreases. By adjusting the pore size of a gel, through manipulating the acrylamide concentration, the resolution of the method can be optimized for a given protein product. Thus, a given gel is physically characterized by its respective composition of acrylamide and bisacrylamide.
In addition to the composition of the gel, the state of the protein is an important component of electrophoretic mobility. In the case of proteins, electrophoretic mobility is dependent on the pK value of the charged groups and the size of the molecule. It is influenced by the type, the concentration, and the pH of the buffer; by the temperature and the field strength; and by the nature of the support material.
Denaturation with Sodium Dodecyl Sulfate
Denaturing PAGE using sodium dodecyl sulfate (SDS) is the most common mode of electrophoresis used in assessing the pharmaceutical quality of protein products. Typically, analytical electrophoresis of proteins is carried out under conditions that ensure dissociation of the proteins into their individual polypeptide subunits and that minimize aggregation of these subunits. The strongly anionic detergent SDS is used in combination with heat to dissociate the proteins before they are loaded on the gel. The denatured polypeptides bind SDS, become negatively charged, and exhibit a consistent charge-to-weight ratio regardless of protein type. Because the amount of SDS bound is almost always proportional to the molecular weight of the polypeptide and is typically independent of its sequence, SDS–polypeptide complexes migrate through polyacrylamide gels in reasonable accordance with the size of the polypeptide.
The electrophoretic mobilities of the resultant detergent–polypeptide complexes all assume the same functional relationship to the molecular weights of the polypeptides. Migration of SDS derivatives is toward the anode in a predictable manner, with low molecular weight complexes migrating faster than larger ones. This means that the molecular weight of a protein can be estimated from its relative mobility in calibrated SDS PAGE and that a single band in such a gel is a criterion of purity.
Modifications to the polypeptide backbone, such as N- or O-linked glycosylation, however, have a significant impact on the apparent molecular weight of a protein. This is due to the fact that SDS does not bind to a carbohydrate moiety in a manner similar to that of the polypeptide. Thus, a consistent charge-to-weight ratio is not maintained. The apparent molecular weight of proteins having undergone post-translational modifications is not a true reflection of the weight of the polypeptide chain.
Reducing Conditions
Polypeptide subunits and their three-dimensional structure can be maintained in proteins by the presence of disulfide bonds. A goal of SDS PAGE analysis under reducing conditions is to disrupt this structure by reducing disulfide bonds. Complete denaturation and dissociation of proteins by treatment with 2-mercaptoethanol or dithiothreitol (DTT) will result in the unfolding of the polypeptide backbone and subsequent complexation with SDS. Under these conditions, the molecular weight of the polypeptide subunits can be calculated by linear regression in the presence of suitable molecular weight standards.
Nonreducing Conditions
For some analyses, complete dissociation of protein to peptide subunits is not desirable. In the absence of treatment with reducing agents, disulfide covalent bonds remain intact, preserving the oligomeric form of the protein. Oligomeric SDS–protein complexes migrate more slowly than their SDS–polypeptide subunits. In addition, nonreduced proteins may not be completely saturated with SDS and hence may not bind the detergent in a constant weight ratio. This makes molecular weight determinations of these molecules less straightforward than analyses of fully denatured polypeptides, because, for valid comparisons, it is necessary that both standards and unknown proteins be in similar configurations. However, the staining of a single band in such a gel is a criterion of purity.
Characteristics of a Discontinuous Buffer System
The most popular electrophoretic method for the characterization of a complex mixture of proteins involves the use of a discontinuous buffer system consisting of two contiguous, but distinct, gels: a resolving or separating (lower) gel and a stacking (upper) gel. The two gels are cast with different porosities, pHs, and ionic strengths. In addition, different mobile ions are used in the gel and electrode buffers. The buffer discontinuity concentrates large volumes of sample in the stacking gel, resulting in improved resolution. When power is applied, a voltage drop develops across the sample solution that drives the proteins into the stacking gel. Glycinate ions from the electrode buffer follow the proteins into the stacking gel. A moving boundary region is rapidly formed with the highly mobile chloride ions in the front and the relatively slow glycinate ions in the rear. A localized high-voltage gradient forms between the leading and trailing ion fronts, causing the SDS–protein complexes to form into a thin zone (stack) and migrate between the chloride and glycinate phases. Within a broad limit, regardless of the height of the applied sample, all SDS proteins condense into a very narrow region and enter the resolving gel as a well-defined, thin zone of high protein density. The large-pore stacking gel does not retard the migration of most proteins and serves mainly as an anticonvective medium. At the interface between the stacking and resolving gels, the proteins experience a sharp retardation due to the restrictive pore size of the resolving gel. Once in the resolving gel, proteins continue to be slowed by the sieving of the matrix. The glycinate ions overtake the proteins, which then move in a space of uniform pH formed by tris(hydroxymethyl)aminomethane (Tris) and glycine. Molecular sieving causes the SDS–polypeptide complexes to separate on the basis of their molecular weights.
Preparation of Gels
In a discontinuous buffer SDS–polyacrylamide gel, it is important to pour the resolving gel, let the gel set, and then pour the stacking gel, because the composition of acrylamide–bisacrylamide, buffer, and pH are different.
Gel Stock Solutions—
30% Acrylamide–Bisacrylamide Solution—
Prepare a solution containing 290 g of acrylamide and 10 g of methylene bisacrylamide per L of warm water, and filter. [note—Acrylamide and methylene bisacrylamide are slowly converted during storage to acrylic acid and bisacrylic acid, respectively. This deamidation reaction is catalyzed by light and alkali. The pH of the solution must be 7.0 or lower. Store the solution in dark bottles at room temperature. Fresh solutions are prepared every month.]
Ammonium Persulfate Solution—
Prepare a small quantity of solution having a concentration of 100 g of ammonium persulfate per L, and store at 4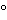 . [note—Ammonium persulfate provides the free radicals that drive polymerization of acrylamide and bisacrylamide. Ammonium persulfate decomposes slowly; therefore, prepare fresh solutions weekly.]
. [note—Ammonium persulfate provides the free radicals that drive polymerization of acrylamide and bisacrylamide. Ammonium persulfate decomposes slowly; therefore, prepare fresh solutions weekly.]
TEMED—
Use an electrophoresis-grade reagent. [note—TEMED accelerates the polymerization of acrylamide and bisacrylamide by catalyzing the formation of free radicals from ammonium persulfate. Because TEMED works only as a free base, polymerization is inhibited at low pH.]
SDS Solution—
Use an electrophoresis-grade reagent. Prepare a solution having a concentration of about 100 g of SDS per L, and store at room temperature.
1.5 M Buffer Solution—
Transfer about 90.8 g of Tris to a 500-mL flask, dissolve in 400 mL of water, adjust with hydrochloric acid to a pH of 8.8, dilute with water to volume, and mix.
1 M Buffer Solution—
Transfer about 60.6 g of Tris to a 500-mL flask, add 400 mL of water, adjust with hydrochloric acid to a pH of 6.8, dilute with water to volume, and mix.
Plate Preparation—
Clean two glass plates (10 cm × 8 cm), the polytef comb, the two spacers, and the silicone rubber tubing (0.6 mm × 35 cm) with mild detergent, rinse thoroughly with water, and blot dry.
Lubricate the spacers and the tubing with nonsilicone grease. Apply the spacers along each of the two short sides of the glass plate 2 mm away from the edges and 2 mm away from the long side corresponding to the bottom of the gel.
Begin to lay the tubing on the glass plate by using one spacer as a guide. Carefully twist the tubing at the bottom of the spacer, and follow the long side of the glass plate. While holding the tubing with one finger along the long side, twist the tubing again, and lay it on the second short side of the glass plate, using the spacer as a guide.
Place the second glass plate in perfect alignment with the first, and hold the gel mold together by hand pressure. Apply two clamps on each of the two short sides of the mold. Carefully apply four clamps on the longer side of the mold, thus forming the bottom of the mold. Verify that the tubing is running along the edge of the glass plates and has not been extruded while placing the clamps. The mold is now ready for the pouring of the gel.
Resolving Gel—
In a conical flask, prepare the appropriate volume of solution, containing the desired concentration of acrylamide, as shown in Table 1. Mix the components in the order shown. Before adding the Ammonium Persulfate Solution and the TEMED, pour the solution into a disposable filtration unit equipped with a nitrocellulose filter having a 0.45-µm porosity, and apply vacuum. Allow the solution to degas by swirling the filtration unit, and disconnect the vacuum when no more bubbles are formed in the solution. Add appropriate amounts of Ammonium Persulfate Solution and TEMED, as shown in Table 1; swirl; and pour immediately into the gap between the two glass plates of the mold. Leave sufficient space for the stacking gel (the length of the teeth of the comb plus 1 cm). Using a pipet, carefully overlay the solution with water-saturated isobutyl alcohol. Leave the gel in a vertical position at room temperature for polymerization.
Table 1. Preparation of Resolving Gel
| Component Volume (mL) per Gel Mold Volume Below | ||||||||
| Solution Component | 5 mL | l0 mL | 15 mL | 20 mL | 25 mL | 30 mL | 40 mL | 50 mL |
| 6% Acrylamide | ||||||||
| 2.6 | 5.3 | 7.9 | 10.6 | 13.2 | 15.9 | 21.2 | 26.5 | |
| 1.0 | 2.0 | 3.0 | 4.0 | 5.0 | 6.0 | 8.0 | 10.0 | |
| 1.3 | 2.5 | 3.8 | 5.0 | 6.3 | 7.5 | 10.0 | 12.5 | |
| 0.05 | 0.1 | 0.15 | 0.2 | 0.25 | 0.3 | 0.4 | 0.5 | |
| 0.05 | 0.1 | 0.15 | 0.2 | 0.25 | 0.3 | 0.4 | 0.5 | |
| 0.004 | 0.008 | 0.012 | 0.016 | 0.02 | 0.024 | 0.032 | 0.04 | |
| 8% Acrylamide | ||||||||
| 2.3 | 4.6 | 6.9 | 9.3 | 11.5 | 13.9 | 18.5 | 23.2 | |
| 1.3 | 2.7 | 4.0 | 5.3 | 6.7 | 8.0 | 10.7 | 13.3 | |
| 1.3 | 2.5 | 3.8 | 5.0 | 6.3 | 7.5 | 10.0 | 12.5 | |
| 0.05 | 0.1 | 0.15 | 0.2 | 0.25 | 0.3 | 0.4 | 0.5 | |
| 0.05 | 0.1 | 0.15 | 0.2 | 0.25 | 0.3 | 0.4 | 0.5 | |
| 0.003 | 0.006 | 0.009 | 0.012 | 0.015 | 0.018 | 0.024 | 0.03 | |
| 10% Acrylamide | ||||||||
| 1.9 | 4.0 | 5.9 | 7.9 | 9.9 | 11.9 | 15.9 | 19.8 | |
| 1.7 | 3.3 | 5.0 | 6.7 | 8.3 | 10.0 | 13.3 | 16.7 | |
| 1.3 | 2.5 | 3.8 | 5.0 | 6.3 | 7.5 | 10.0 | 12.5 | |
| 0.05 | 0.1 | 0.15 | 0.2 | 0.25 | 0.3 | 0.4 | 0.5 | |
| 0.05 | 0.1 | 0.15 | 0.2 | 0.25 | 0.3 | 0.4 | 0.5 | |
| 0.002 | 0.004 | 0.006 | 0.008 | 0.01 | 0.012 | 0.016 | 0.02 | |
| 12% Acrylamide | ||||||||
| 1.6 | 3.3 | 4.9 | 6.6 | 8.2 | 9.9 | 13.2 | 16.5 | |
| 2.0 | 4.0 | 6.0 | 8.0 | 10.0 | 12.0 | 16.0 | 20.0 | |
| 1.3 | 2.5 | 3.8 | 5.0 | 6.3 | 7.5 | 10.0 | 12.5 | |
| 0.05 | 0.1 | 0.15 | 0.2 | 0.25 | 0.3 | 0.4 | 0.5 | |
| 0.05 | 0.1 | 0.15 | 0.2 | 0.25 | 0.3 | 0.4 | 0.5 | |
| 0.002 | 0.004 | 0.006 | 0.008 | 0.01 | 0.012 | 0.016 | 0.02 | |
| 14% Acrylamide | ||||||||
| 1.4 | 2.7 | 3.9 | 5.3 | 6.6 | 8.0 | 10.6 | 13.8 | |
| 2.3 | 4.6 | 7.0 | 9.3 | 11.6 | 13.9 | 18.6 | 23.2 | |
| 1.2 | 2.5 | 3.6 | 5.0 | 6.3 | 7.5 | 10.0 | 12.5 | |
| 0.05 | 0.1 | 0.15 | 0.2 | 0.25 | 0.3 | 0.4 | 0.5 | |
| 0.05 | 0.1 | 0.15 | 0.2 | 0.25 | 0.3 | 0.4 | 0.5 | |
| 0.002 | 0.004 | 0.006 | 0.008 | 0.01 | 0.012 | 0.016 | 0.02 | |
| 15% Acrylamide | ||||||||
| 1.1 | 2.3 | 3.4 | 4.6 | 5.7 | 6.9 | 9.2 | 11.5 | |
| 2.5 | 5.0 | 7.5 | 10.0 | 12.5 | 15.0 | 20.0 | 25.0 | |
| 1.3 | 2.5 | 3.8 | 5.0 | 6.3 | 7.5 | 10.0 | 12.5 | |
| 0.05 | 0.1 | 0.15 | 0.2 | 0.25 | 0.3 | 0.4 | 0.5 | |
| 0.05 | 0.1 | 0.15 | 0.2 | 0.25 | 0.3 | 0.4 | 0.5 | |
| 0.002 | 0.004 | 0.006 | 0.008 | 0.01 | 0.012 | 0.016 | 0.02 | |
After polymerization is complete (about 30 minutes later), pour off the overlay, and wash the top of the gel several times with water to remove the isobutyl alcohol overlay and any unpolymerized acrylamide. Drain as much fluid as possible from the top of the gel, then remove any remaining water with the edge of a paper towel.
Stacking Gel—
In a conical flask, prepare the appropriate volume of solution containing the desired concentration of acrylamide, as shown in Table 2.
Mix the components in the order shown. Before adding the Ammonium Persulfate Solution and the TEMED, pour the solution into a disposable filtration unit equipped with a nitrocellulose filter having a 0.45-µm porosity, and apply vacuum. Allow the solution to degas by swirling the filtration unit, and disconnect the vacuum when no more bubbles are formed in the solution. Add appropriate amounts of Ammonium Persulfate Solution and TEMED as shown in Table 2, swirl, and pour immediately into the gap between the two glass plates of the mold directly onto the surface of the polymerized Resolving Gel. Immediately insert a clean polytef comb into the stacking gel solution, being careful to avoid trapping air bubbles. Add more stacking gel solution to fill the spaces of the comb completely. Leave the gel in a vertical position, and allow it to polymerize at room temperature. After polymerization is complete (about 30 minutes later), carefully remove the polytef comb, and proceed as directed below.
Table 2. Preparation of Stacking Gel
| Component Volume (mL) per Gel Mold Volume Below | ||||||||
| Solution Component | 1 mL | 2 mL | 3 mL | 4 mL | 5 mL | 6 mL | 8 mL | 10 mL |
| Water | 0.68 | 1.4 | 2.1 | 2.7 | 3.4 | 4.1 | 5.5 | 6.8 |
| 30% Acrylamide–Bisacrylamide Solution | 0.17 | 0.33 | 0.5 | 0.67 | 0.83 | 1.0 | 1.3 | 1.7 |
| 1.0 M Buffer Solution | 0.13 | 0.25 | 0.38 | 0.5 | 0.63 | 0.75 | 1.0 | 1.25 |
| SDS Solution | 0.01 | 0.02 | 0.03 | 0.04 | 0.05 | 0.06 | 0.08 | 0.1 |
| Ammonium Persulfate Solution | 0.01 | 0.02 | 0.03 | 0.04 | 0.05 | 0.06 | 0.08 | 0.1 |
| TEMED | 0.001 | 0.002 | 0.003 | 0.004 | 0.005 | 0.006 | 0.008 | 0.01 |
Electrophoretic Separation
Sample Buffer 1—
Dissolve 1.89 g of Tris, 5.0 g of SDS, 50 mg of bromophenol blue, and 25.0 mL of glycerol in 100 mL of water. Adjust with hydrochloric acid to a pH of 6.8, and dilute with water to 125 mL. Before use, dilute with an equal volume of water or sample, and mix.
Sample Buffer 2 (for reducing conditions)—
Prepare as directed in Sample Buffer 1 except to add 12.5 mL of 2-mercaptoethanol before adjusting the pH. Alternatively, prepare as directed for Sample Buffer 1 except to start with about 1.93 g of Tris and add a suitable quantity of DTT to obtain a final 100 µM DTT concentration.
Running Buffer—
Dissolve 151.4 g of Tris, 721.0 g of aminoacetic acid (glycine), and 50.0 g of SDS in water; dilute with water to 5000 mL; and mix to obtain a stock solution. Immediately before use, dilute this stock solution with water to 10 times its volume, mix, and adjust to a pH between 8.1 and 8.8.
Procedure—
Rinse the wells immediately with water or with the Running Buffer to remove any unpolymerized acrylamide. (If necessary, straighten the teeth of the Stacking Gel with a blunt hypodermic needle attached to a syringe.) Remove the clamps on one short side, carefully pull out the tubing, and replace the clamps. Proceed similarly on the other short side. Remove the tubing from the bottom part of the gel.
Mount the completed gel in the electrophoresis apparatus. Add the electrophoresis buffers to the top and bottom reservoirs. Remove any bubbles that become trapped at the bottom of the gel between the glass plates. [note—Removal is best done with a bent hypodermic needle attached to a syringe. Never prerun the gel before loading the samples, because that will destroy the discontinuity of the buffer systems. Before loading the sample, carefully rinse the slot with Running Buffer.]
Prepare the test and standard solutions in the recommended Sample Buffer, and treat as directed in the individual monograph. Apply the appropriate volume of each solution to the Stacking Gel wells.
Start the electrophoresis under the conditions recommended by the manufacturer of the equipment. Electrophoresis running time and current or voltage may need to be varied in order to achieve optimum separation. Check that the dye front is moving into the Resolving Gel. When the dye has reached the bottom of the gel, stop the electrophoresis. Remove the gel assembly from the apparatus, and separate the glass plates. Remove the spacers, cut off and discard the Stacking Gel, and immediately proceed with staining.
Detection of Proteins in Gels
Coomassie staining is the most common protein staining method, with a detection level on the order of 1 to 10 µg of protein per band. Silver staining is the most sensitive method for staining proteins in gels, because a band containing 10 to 100 ng can be detected; but the method is more cumbersome and less rugged. All of the steps in gel staining are performed at room temperature with gentle agitation (e.g., on a rocking platform shaker or equivalent). Gloves must be worn when staining the gels to prevent fingerprint residue staining.
Reagents—
Coomassie Staining Solution—
Prepare a solution of Coomassie brilliant blue R-250 having a concentration of 1.25 g per L in a mixture of water, methanol, and glacial acetic acid (5:4:1). Filter, and store at room temperature.
Destaining Solution—
Prepare a mixture of water, methanol, and glacial acetic acid (5:4:1).
Fixing Solution 1—
Prepare a mixture of water, methanol, and trichloroacetic acid (5:4:1).
Fixing Solution 2—
Transfer 250 mL of methanol to a 500-mL volumetric flask, add 0.27 mL of formaldehyde, dilute with water to volume, and mix.
Silver Nitrate Reagent—
To a mixture of 40 mL of 1 M sodium hydroxide and 3 mL of ammonium hydroxide, add, dropwise and with stirring, 8 mL of a 200 g per L solution of silver nitrate; dilute with water to 200 mL, and mix.
Developing Solution—
Transfer 2.5 mL of a citric acid solution (2 in 100) and 0.27 mL of formaldehyde to a 500.0-mL volumetric flask, dilute with water to volume, and mix.
Stopping Solution—
Prepare a 10% (v/v) solution of acetic acid.
Coomassie Staining—
Immerse the gel in an excess of Coomassie Staining Solution, and incubate for at least 1 hour. Remove the Coomassie Staining Solution. Destain the gel with an excess of Destaining Solution. Change the Destaining Solution several times, until the stained protein bands are clearly distinguishable on a clear background. The more thoroughly the gel is destained, the smaller the amount of protein that can be detected. Destaining can be accelerated by including a few g of anion-exchange resin or a small sponge in the Destaining Solution. [note—The acid–alcohol solutions used in this procedure do not completely fix proteins in the gel. This can lead to losses of some low molecular weight proteins during the staining and destaining of thin gels. Permanent fixation is obtainable by incubating the gel in Fixing Solution 1 for 1 hour before it is immersed in the Coomassie Staining Solution.]
Silver Staining—
Immerse the gel in an excess of Fixing Solution 2, and incubate for 1 hour. Remove Fixing Solution 2, add fresh Fixing Solution 2, and incubate for at least 1 hour, or overnight if convenient. Discard Fixing Solution 2, and wash the gel in an excess of water for 1 hour. Soak the gel for 15 minutes in a 1% solution of glutaraldehyde (v/v). Wash the gel twice, for 15 minutes each time, with an excess of water. Soak the gel in fresh Silver Nitrate Reagent for 15 minutes in darkness. Wash the gel three times, for 5 minutes each time, with an excess of water. Immerse the gel for about 1 minute in Developing Solution until satisfactory staining has been obtained. Stop the development by incubation in the Stopping Solution for 15 minutes, then rinse the gel with water, and proceed with drying as indicated below.
Drying of Gels
For Coomassie staining, after the destaining step, incubate the gel in a glycerol solution (1 in 10) for at least 2 hours. For silver staining, add to the final rinsing step a 5-minute incubation in a glycerol solution (1 in 50).
Immerse two sheets of porous cellophane in water, and incubate for 5 to 10 minutes. Place one of the sheets on a drying frame. Carefully lift the gel, and place it on the cellophane sheet. Remove any trapped air bubbles, and pour a few mL of water around the edges of the gel. Place the second sheet on top, and remove any trapped air bubbles. Complete the assembly of the drying frame. Place in a drying oven, leave at room temperature until dry, or use a commercial gel dryer.
Molecular Weight Determination
Molecular weights of proteins are determined by comparison of their mobilities with those of several marker proteins of known molecular weight. Mixtures of proteins with precisely known molecular weights blended for uniform staining are available for calibrating gels. They are available in various molecular weight ranges. Concentrated stock solutions of proteins of known molecular weight are diluted in a sample buffer and loaded on the same gel as the protein sample to be tested.
Immediately after the gel has been run, the position of the bromophenol blue tracking dye is marked to identify the leading edge of the electrophoretic ion front. This can be done by cutting notches in the edges of the gel or by inserting a needle soaked in India ink into the gel at the dye front. After staining, measure the migration distances of each protein band (markers and unknowns) from the top of the Resolving Gel. Divide the migration distance of each protein by the distance traveled by the tracking dye. The normalized migration distances so obtained are called the relative mobilities of the proteins (relative to the dye front) and conventionally denoted as RF . Construct a (semilogarithmic) plot of the logarithm of the molecular weights (MR) of the protein standards as functions of the RF values. [note—The graphs are slightly sigmoid.] From the graph so obtained, estimate the unknown molecular weights by linear regression analysis or interpolation, as long as unknown samples are positioned along the linear part of the graph.
If the proteins of the molecular weight marker are not distributed along 80% of the length of the gel and over the required separation range (i.e., the range covering the product and its dimer or the products and its related impurities), and the separation obtained for the relevant protein bands does not show a linear relationship between the logarithm of the molecular weight and the RF , the test is not valid.
Quantification of Impurities
Where the impurity limit is specified in the individual monograph, a reference solution corresponding to that level of impurity is prepared by diluting the test solution. For example, where the limit is 5.0%, a reference solution is a 1 in 20 dilution of the test solution. No impurity—any band other than the main band—in the electrophoretogram obtained from the test solution is more intense than the main band obtained with the reference solution.
Under validated conditions and when using the Coomassie staining procedure, impurities may be quantified by normalization to the main band, using an integrating densitometer. In this case, the responses must be validated for linearity.
Auxiliary Information— Please check for your question in the FAQs before contacting USP.
| Topic/Question | Contact | Expert Committee |
| General Chapter | Alexey Khrenov, Ph.D
Senior Scientific Associate 1-301-816-8345 |
(BBPP05) Biologics and Biotechnology - Proteins and Polysaccharides |
USP32–NF27 Page 500
Pharmacopeial Forum: Volume No. 32(2) Page 580